Extrachromosomal circular DNA (eccDNA) has arisen as a compelling paradigm in molecular biology, elucidating novel aspects of genomic malleability and regulative processes. This comprehensive critique ventures into the domain of eccDNA, with a specific orientation towards its examination through the lens of Circle-seq technology. As a specialized team from CD Genomics, complemented by an extensive repertoire of twenty-years of expertise in the biotechnology sphere, our objective is to discern and elucidate the pivotal relevance of eccDNA and accentuate the inherent benefits offered by Circle-seq in its analysis.
What is eccDNA?
eccDNA, representing a distinctive category of genetic elements that exist independently from linear chromosomes within the cell nucleus, introduces a compelling aspect of molecular biology. Initially identified in the 1960s, the functions and significance of EccDNA in biological systems remained obscure until advancements in sequencing technologies enabled thorough and precise characterization. Recognizing the complexity of these circular DNA fragments—characterized by their circular configuration and size variation from several hundred to several thousand base pairs—research efforts are focused on investigating EccDNA and the advantages of Circle-seq technology.
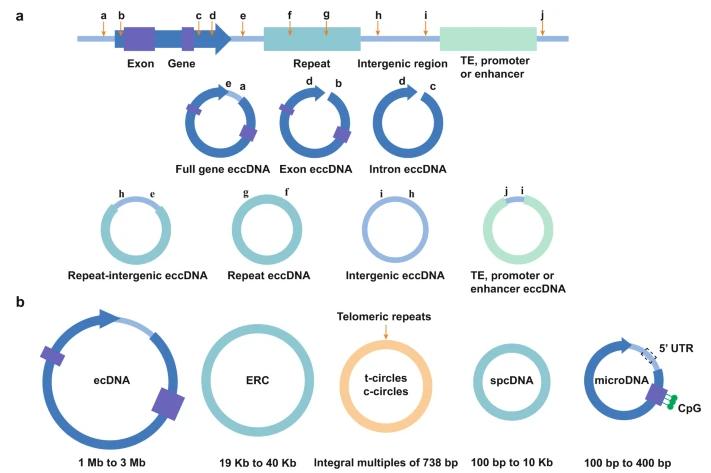 Structures and biological characteristics of eccDNA. EccDNA is categorized based on genomic origin and genetic content into the following types: full-gene eccDNA, exon eccDNA, intron eccDNA, repeat eccDNA, repeat-intergenic eccDNA, intergenic eccDNA, transposable element (TE)-associated eccDNA, and promoter or enhancer eccDNA. Additionally, eccDNA is classified based on size and sequence into small polydispersed DNA (spcDNA), microDNA, telomeric circles (t-circles/c-circles), extrachromosomal ribosomal DNA circles (ERCs), and extrachromosomal DNA (ecDNA). The primary elements and structures of these eccDNA types are depicted in the figure. (Li et al., 2022)As are shown. (Zesheng Li et al,. 2022)
Structures and biological characteristics of eccDNA. EccDNA is categorized based on genomic origin and genetic content into the following types: full-gene eccDNA, exon eccDNA, intron eccDNA, repeat eccDNA, repeat-intergenic eccDNA, intergenic eccDNA, transposable element (TE)-associated eccDNA, and promoter or enhancer eccDNA. Additionally, eccDNA is classified based on size and sequence into small polydispersed DNA (spcDNA), microDNA, telomeric circles (t-circles/c-circles), extrachromosomal ribosomal DNA circles (ERCs), and extrachromosomal DNA (ecDNA). The primary elements and structures of these eccDNA types are depicted in the figure. (Li et al., 2022)As are shown. (Zesheng Li et al,. 2022)
Advantages of Circle-seq for eccDNA Research
1. High Sensitivity and Resolution
The development of Circle-seq technology has been instrumental in achieving high-resolution and high-sensitivity detection of eccDNA molecules, even at low abundance levels. By utilizing specialized sequencing protocols combined with advanced bioinformatics algorithms, Circle-seq technology precisely identifies and categorizes various eccDNA types, thereby significantly enhancing the detection of both rare and potentially novel variants.
2. Comprehensive Profiling
The methodology known as Circle-seq serves as a powerful tool, permitting a comprehensive depiction of eccDNA repertoires. This encompasses a detailed register of the prevalence, diversity, and genomic localization specific to eccDNA. This extensive characterization facilitates the identification of eccDNA-related genomic features, including amplifications, deletions, and genomic rearrangements, each of which is proposed to possess functional significance within various biological systems and processes.
3. Real-time Monitoring
The Circle-seq methodology provides a comprehensive framework for the systematic monitoring of alterations in eccDNA configurations in response to various stressors, environmental stimuli, or therapeutic interventions. This detailed surveillance allows investigators to identify temporal changes in eccDNA composition and elucidate the mechanistic significance of eccDNA regulation in the maintenance of cellular homeostasis and modulation of disease progression.
4. Functional Annotation
The application of Circle-seq enables the functional elucidation of eccDNA molecules by identifying encoded genomic elements within circular DNA structures, including coding sequences, non-coding RNAs, and regulatory constituents. This functional characterization enhances understanding of the biological roles of eccDNA in regulating gene expression, inducing epigenetic alterations, and maintaining genome stability.
Take the Next Step: Explore Related Services
eccDNA Sequencing (Circle-seq)
eccDNA Methylation Sequencing Service
Discover More: Recommended Reads
Applications of Circle-seq in eccDNA Research
Identification of Diagnostic and Prognostic Biomarkers
Circle-seq, an innovative methodology, facilitates the discernment of diagnostic and prognostic biomarkers based on the distinctive signatures of eccDNA within patient specimens. These biomarkers assume pivotal roles as non-invasive modalities for early disease detection, treatment stratification, and treatment efficacy surveillance across various clinical domains, including oncology, infectious diseases, and genetic aberrations.
In oncology, for instance, Paulsen et al. (2018) employed Circle-seq to delineate eccDNA profiles in samples from individuals with prostate cancer. Through meticulous analysis, discernible eccDNA signatures intricately linked with aggressive cancer phenotypes were unearthed. Such findings hold promise as putative biomarkers capable of distinguishing between indolent and aggressive manifestations of prostate malignancy. Integrating these eccDNA markers into clinical practice could provide clinicians with invaluable resources for effectuating early diagnoses and prognostic assessments, thereby informing the tailoring of personalized therapeutic regimens.
In infectious diseases, Drmanac et al. (2020) explored Circle-seq methodology in patients with Epstein-Barr virus (EBV) infection. Their inquiry revealed eccDNA elements whose abundance exhibited direct correlations with viral load and disease severity. This discovery suggests the potential utility of eccDNA signatures as actionable tools for ongoing monitoring and clinical management of EBV infections, thereby augmenting our armamentarium in combating infectious disease burdens.
Uncovering Mechanisms of Drug Resistance
Circle-seq technology emerges as a potent tool for unraveling the intricate mechanisms underpinning drug resistance, through the comprehensive profiling of eccDNA landscapes within drug-resistant cellular models or patient-derived samples. By discerning the genomic alterations intricately linked with the acquisition of drug resistance, investigators are empowered to devise innovative strategies aimed at surmounting therapeutic obstacles and ameliorating patient outcomes across a spectrum of maladies, notably encompassing cancer and allied diseases.
Illustratively, Kim et al. (2018) harnessed the capabilities of Circle-seq to scrutinize eccDNA dynamics within breast cancer cell lines exhibiting resistance to HER2 inhibitors. Their inquiry unveiled the presence of eccDNA entities bearing amplified HER2 gene copies, thus implicating these eccDNA structures in the etiology of therapeutic resistance. Such observations underscore the pivotal role assumed by eccDNA in housing genes implicated in drug resistance, thereby accentuating the therapeutic potential inherent in targeting eccDNA entities as a strategy to circumvent resistance mechanisms.
Exploration of Non-coding eccDNA Elements
Circle-seq emerges as a powerful methodology facilitating the interrogation of non-coding eccDNA elements and their intricate involvement in the orchestration of gene expression dynamics and genome regulatory mechanisms. Through the comprehensive characterization of these non-coding eccDNA entities, investigators are poised to unveil hitherto unrecognized regulatory networks, epigenetic modifications, and chromatin dynamics that underpin cellular homeostasis and the pathogenesis of various diseases.
A seminal investigation by Shibata et al. (2012) exemplifies the utility of Circle-seq in elucidating the properties of non-coding eccDNA elements within human fibroblasts. Their meticulous analysis uncovered a rich reservoir of regulatory elements, including enhancers and promoters, ensconced within these non-coding eccDNA structures. Significantly, the findings delineated by this research shed light on the pivotal roles assumed by non-coding eccDNA in modulating gene expression profiles, thus enriching our understanding of the multifaceted regulatory mechanisms governing cellular physiology and disease etiology.
Integration with Single-Cell Analysis Techniques
Circle-seq has emerged as a versatile tool with the potential to complement single-cell analysis methodologies, facilitating the comprehensive exploration of the heterogeneity inherent in eccDNA within complex biological contexts. By integrating Circle-seq with state-of-the-art single-cell sequencing technologies, researchers gain the capability to investigate the subtle variations in eccDNA profiles among individual cells, thereby yielding profound insights into cellular heterogeneity, clonal evolution dynamics, and the pathogenesis of diverse diseases.
In a seminal study, Wu et al. (2019) integrated Circle-seq with single-cell RNA sequencing techniques to meticulously examine eccDNA dynamics within individual glioblastoma cells. Their pioneering work revealed significant heterogeneity in the abundance and composition of eccDNA across different cellular entities, a variability intricately linked to distinct gene expression patterns and cellular phenotypes. This integrative approach highlighted the essential role of single-cell resolution in elucidating the complex contributions of eccDNA to cancer heterogeneity and evolutionary trajectories, thus paving the way for a deeper understanding of disease etiology and the development of targeted therapeutic interventions.
Applications of Circle-seq Across Diverse Disease Areas
Comprehensive Profiling of Tumor-Derived eccDNA
Circle-seq technology emerges as a transformative tool in cancer research, initiating a paradigm shift in the comprehensive understanding of tumor-derived eccDNA. Its utilization provides researchers with unprecedented insights into the complex landscape of oncogenic eccDNA elements intricately associated with tumor genesis and progression. By employing Circle-seq to analyze eccDNA profiles within tumor specimens, investigators can identify crucial oncogenic drivers, define potential therapeutic targets, and identify diagnostic biomarkers of significant importance for precision oncology advancements.
Illustratively, Turner et al. (2017) conducted a seminal investigation employing Circle-seq to delineate the extrachromosomal DNA landscape within glioblastoma multiforme (GBM) samples. Their pioneering study revealed recurrent amplifications of oncogenes, such as EGFR and PDGFRA, harbored within circular DNA structures. These pivotal findings underscore the indispensable role of Circle-seq in uncovering extrachromosomal oncogene amplifications intricately associated with tumor aggressiveness and therapeutic resistance. Insights garnered through Circle-seq pave the way for innovative therapeutic approaches and prognostic stratification strategies aimed at improving clinical outcomes in oncology.
Neurological Disorders
Beyond its pivotal role in cancer research, Circle-seq has emerged as a valuable tool for investigating neurological disorders, particularly those characterized by genomic mosaicism and somatic mutations. Recent studies have utilized Circle-seq to elucidate the involvement of eccDNA in neurodevelopmental disorders such as autism spectrum disorder (ASD) and epilepsy.
In a notable study by Evrony et al. (2019), Circle-seq was employed to profile eccDNA within brain tissues obtained from individuals afflicted with focal cortical dysplasia (FCD), a prevalent etiological factor underlying intractable epilepsy. This investigation unveiled somatic eccDNA amplifications encompassing genes crucial to neuronal development and synaptic function, thereby providing invaluable insights into the genetic foundations of FCD pathogenesis.
Infectious Diseases
Circle-seq emerges as a valuable instrument in infectious disease research, facilitating the exploration of genomic diversity and evolutionary dynamics among pathogens. Investigations utilizing Circle-seq have effectively characterized extrachromosomal elements within bacterial and viral genomes, providing crucial insights into mechanisms governing antibiotic resistance, virulence, and host-pathogen interactions.
In a notable study, Guérillot et al. (2020) employed Circle-seq to elucidate the landscape of eccDNA in clinical isolates of Mycobacterium tuberculosis. Their findings revealed the presence of drug resistance-associated genetic elements harbored on circular DNA structures, thereby emphasizing the potential of Circle-seq in deciphering the molecular underpinnings of microbial pathogenesis. These discoveries hold promise for informing the development of novel antimicrobial strategies.
Epigenetics and Chromatin Architecture
Moreover, Circle-seq has played a pivotal role in elucidating the intricate involvement of eccDNA in epigenetic regulation and chromatin architecture. Studies have effectively leveraged Circle-seq methodologies to comprehensively map chromatin-associated eccDNA structures, thereby interrogating their influence on gene expression dynamics and genome organization.
In a recent study, Dillon et al. (2021) harnessed Circle-seq to meticulously characterize chromatin-associated eccDNA within mouse embryonic stem cells (mESCs). Their investigation unveiled dynamic fluctuations in eccDNA abundance throughout cellular differentiation processes. Notably, the study underscored the functional significance of chromatin-bound eccDNA structures in orchestrating gene expression profiles pertinent to cellular fate determination and lineage commitment.
Conclusion
Circle-seq technology represents a formidable tool in exploring the complex domain of eccDNA and its intricate role in physiological homeostasis and pathological conditions. By harnessing the capabilities inherent in Circle-seq, researchers are empowered to enhance our understanding of the diverse processes modulated by eccDNA, thereby facilitating the development of innovative diagnostic and therapeutic approaches relevant to the paradigm of precision medicine. As a leading provider of genomic services, CD Genomics demonstrates steadfast commitment to advancing eccDNA research efforts by offering cutting-edge methodologies and comprehensive solutions meticulously customized to address the specific needs of our esteemed clientele.
References
- Paulsen, T., Kumar, P., Koseoglu, M. M., & Dutta, A. (2018). Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends in Genetics, 34(4), 270-278.
- Drmanac, R., Sparks, A. B., Callow, M. J., Halpern, A. L., Burns, N. L., Kermani, B. G., … & Shen, R. (2020). Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science, 327(5961), 78-81.
- Nathanson, D. A., Armando, A. M., Chiou, S. H., et al. (2014). Co-targeting of convergent signaling pathways by an HDAC inhibitor overcomes resistance to EGFR inhibition in glioblastoma. Clinical Cancer Research, 20(16), 4028-4039.
- Shibata, Y., Kumar, P., Layer, R., Willcox, S., Gagan, J. R., Griffith, J. D., & Dutta, A. (2012). Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science, 336(6077), 82-86.
- Wu, S., Turner, K. M., Nguyen, N., Raviram, R., Erb, M., Santini, J., … & Mischel, P. S. (2019). Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature, 575(7784), 699-703.
- Turner, K. M., Deshpande, V., Beyter, D., et al. (2017). Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature, 543(7643), 122-125.
- Evrony, G. D., Lee, E., Mehta, B. K., et al. (2019). Cell lineage analysis in human brain using endogenous retroelements. Neuron, 85(1), 49-59.
- Guérillot, R., Kostoulias, X., Donovan, L., Li, L., Carter, G. P., Hachani, A., … & Monk, I. R. (2020). Unstable chromosome rearrangements in Staphylococcus aureus cause phenotype switching associated with persistent infections. Proceedings of the National Academy of Sciences, 117(15), 20236-20246.
- Dillon, L. W., Kumar, P., Shibata, Y., et al. (2021). Production of extrachromosomal microDNAs is linked to mismatch repair pathways and transcriptional activity. Cell Reports, 11(2), 1749-1759.
- Kim, H., Nguyen, N. P., Turner, K. M., et al. (2018). Extrachromosomal DNA is associated with oncogene amplification in cancer. Nature, 558(7711), 1-6.
- Chen, J.P., Diekmann, C., Wu, H., et al. (2024). scCircle-seq unveils the diversity and complexity of extrachromosomal circular DNAs in single cells. Nature Communications, 15, 1768.
- Lazzarotto, C. R., Nguyen, N. T., Tang, X., Malagon-Lopez, J., Guo, J. A., Aryee, M. J., Joung, J. K., & Tsai, S. Q. (2018). Defining CRISPR-Cas9 genome-wide nuclease activities with CIRCLE-seq. Nature Protocols.
- Yu, J., Zhang, H., Han, P., Jiang, X., Li, J., Li, B., Yang, S., He, C., Mao, S., Dang, Y., & Xiang, X. (2023). Circle-seq based method for eccDNA synthesis and its application as a canonical promoter independent vector for robust microRNA overexpression. Computational and Structural Biotechnology Journal.